Microscopy Nodes
Microscopy in Blender
Microscopy Nodes is a Blender add-on for visualizing high-dimensional microscopy data—designed for scientists, or anyone working with biological images 😊.
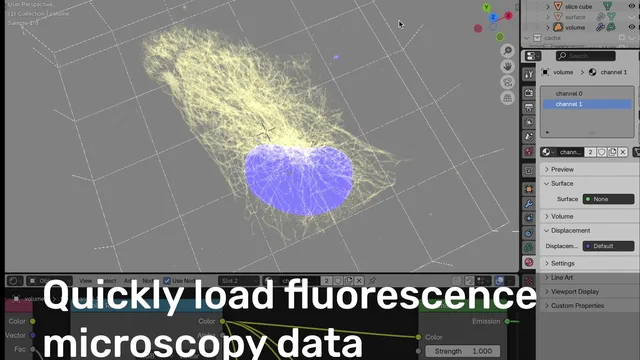
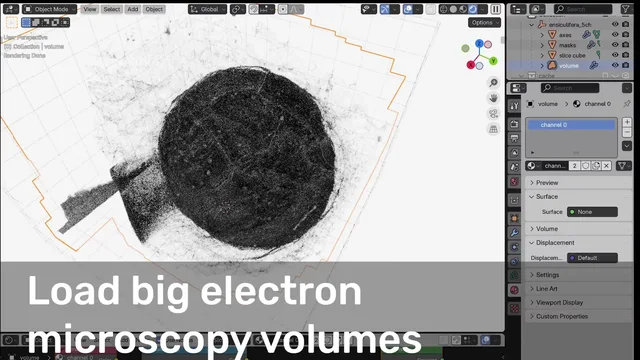
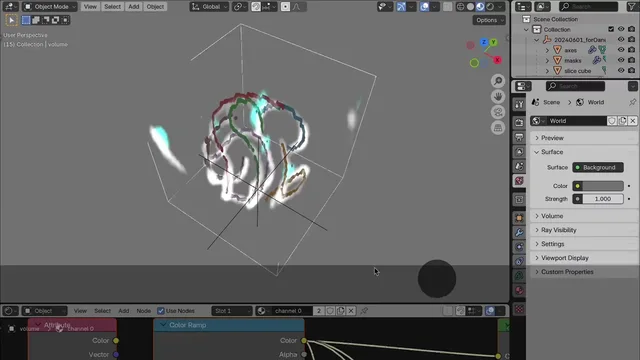
For any type of microscopy: fluorescence, electron microscopy, or anything in between! This tool helps you turn complex 3D+ datasets into stunning, accurate, and animatable visualizations.
Usage questions are mainly answered on the image.sc forum
What It Does
Microscopy Nodes supports importing up to 5D microscopy datasets (XYZ + time + channels) from .tif and OME-Zarr files, setting easy and adaptable settings to start with visualizing your data.
| Feature | Description |
|---|---|
| 5D Support | Load .tif and .zarr files with any axis order ('tzcyx' or any subset) |
| Channel Interface | Define how to load each channel: volume, surface, label mask |
| Colors and LUTs | Easy picking of colors per channel or non-linear LUT selection from many colormaps. |
| Intuitive Slicing | Slice any object by moving the Slicing Cube, as you would move any other Blender object |
| Scales | 3D scale grid for accurate representation and physical Blender scales for easy registration. |
| Large Volumes | Build your animation and visualization on a downscaled version, render with your massive dataset! |
Video tutorials
Check out the video tutorial playlist on YouTube for quick guides on:
- Installation
- Loading data
- Fluorescence & EM visualization
- Making presentation-ready renders
youtube videos are and tutorials are now slightly outdated but will be updated within May/June 2025

First use
- Load your file (local path or URL) into the Microscopy Nodes panel in Scene Properties
- The metadata will auto-load, and you can define how each channel is visualized
- Adjust per-channel options like:
- Volume or isosurface rendering
- Label masks
- Emission, resolution, and colors
- Customize dataset settings like:
- Axis order
- Physical pixel size
- Reload behavior & storage location
More detail in the full docs.
Show Off Your Visualizations!
If you create something cool using Microscopy Nodes, share it!
Tag me @GrosOane on Bluesky or use the hashtag #microscopynodes.
If you publish with this add-on, please cite the preprint:
Interface
The Microscopy Nodes panel can be found in Scene Properties:

Load any tif or zarr file by inputting the path or URL in the appropriate window in the Microscopy Nodes panel. This will read out metadata and prompt you to define how you want to load the data.
What's New
2.2.0 May 16th, 2025
Implements some fun new features that make playing with colors and intensities much easier, fixes bugs. Written and video tutorials for this version are coming in May/June 2025 (soon).
Implements:
- new color selection in channel list
- new setup for shader nodes (splits intensity and color more cleanly)
- Adds color LUTs to all objects, with under right-click on the ramp a lot of selection of premade colormaps from the cmap library
- Preference settings under Edit > Preferences > Add-Ons > Microscopy Nodes, allows you to set the default options of the Microscopy Nodes panels
- Adds a downscaled option for .tif data to load if the data is over 4 GiB
Fixes:
- floating point data should now work
- 32bit data should now work
- rescaling between different sizes of the data and physical spaces is more clean